

Site Directed Mutagenesis-Primer Selection and Protocols
 |
 |
Quick change is the fastest way to create a Amino Acid change in your protein. It works directly on circular plasmid targets with size not bigger then 10kb. It is good idea to move your target DNA sequence to a small vector before to do SDM.
How to select primers
Paste your Nucleotide sequence below. You can translate it to Amino Acid sequence. Find your Amino Acid you want to change (Your browser Edit-Find and type Amino Acids stretch with one underbarars on both sides (_A__G_)). Change the nucleotide sequence. Select a primer comprising the change amino acid. Change the selected oligo to Reverse - Complimentary in the form below. What you must have at the end is a primer dimmer (two overlapping primers).
Considerations for the primer selection are:
♦ Both of the mutagenic primers must contain the desired mutation and anneal to the same sequence on opposite strands of the plasmid.
♦ Primers should be between 25 and 45 bases in length, with a melting temperature (Tm) of ≥78°C. Primers longer than 45 bases may be used, but using longer primers increases the likelihood of secondary structure formation, which may affect the efficiency of the mutagenesis
reaction.
♦ The desired mutation (deletion or insertion) should be in the middle of the primer with ~10–15 bases of correct sequence on both sides.
How does it work:
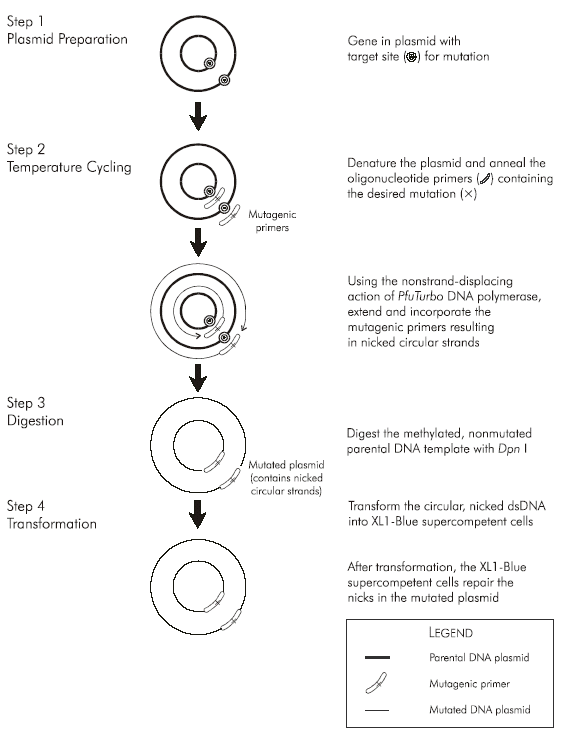
Plasmid purification for Quick Change is the very critical step. If you break or nick your DNA it will effect the outcome of the SDM. Use a kit for plasmid purification. Preferably use cesioum preps.
Pfu polymerase - I try many types of proof reading polymerases, still pfu is the best one for SDM-Quick change.
DpnI restriction enzyme- The Dpn I endonuclease (target sequence: 5´-Gm6ATC-3´) is specific for methylated and hemimethylated DNA and is used to digest the parental DNA template and to select for mutation-containing synthesized DNA. DNA isolated from almost all E. coli strains is dam methylated and therefore susceptible to Dpn I digestion. Any way check the strain of your E. coli stock. If you are not sure what the strain your plasmid comes from use DH5A or Top 10 competent cells and transform your DNA, then you extract new DNA for site directed mutagenesis.
Step1
Set a PCR
Although you may very your PCR conditions, start from somewhere...
50µl PCR
50-100ng dsDNA (Plasmid template-amount is preferably as low as possible. It should be a faint band on 1% Agarose gel)
1µl (10mM dNTPs) (Excess of dNTPs)
6 pmol Forward primer (Excess of primer)
6 pmol Reverse primer (Excess of primer)
5 µl of 10× reaction buffer (Comes with the polymerase)
1 µl of PfuTurbo DNA polymerase (2.5 U/µl)
double-distilled water (ddH2O) to a final volume of 50 µl
Cycling Parameters for the QuikChange Site-Directed
95°C 30 seconds
(95°C 30 seconds, 55°C 1 minute, 68°C 10 minutes ) x 20 ~ 30 cycles
Step 2
Add 1µl Dpn 1(NEB) directly into the PCR mix and digest for 1h @ 37°C (preferably in water bath).
Step 3
Transform 5~10 µl of the PCR mix into realy good competent cells.